It is a bit frustrating because other parts of my life have been interfering with my studio time! This despite the fact that I am “retired” from Georgetown. Retired is in quotes because a fellow faculty member really needs my expertise in image analysis. IA, as we like to abbreviate image analysis, is the use of computers to analyze and quantify features of images acquired using microscopes. In my case, it is a fluorescence confocal microscope. Here are some impressive images as shown on the Leica website (scroll down!!).
Although each layer of a confocal 3D image is monochrome (black and white), the composite image is in RBG, that is, red, blue, green: the appropriate display for computer screens. Advances in computer technology and advances in chemistry of fluorophores and the molecular biology of natural proteins from jellyfish that are fluorescent have revolutionized imaging during my professional career. Totally amazing.
The current project that requires IA is a study to develop a method to measure the effects of naturally-derived compounds (for example, as found in green tea) on DNA damage that occurs in cell DNA during cancer progression. The team at Georgetown directed by Fung-Lung Chung has found a way to identify DNA damage in images of cells. What they need help on is automating the analysis to measure DNA damage in individual cells stained using three colors. That’s about all I want to say about that, but IA can be really really fun to do and develop, and really really annoying. Kind of like writing software if any of you all out there have tried it. It only takes one mistake to mess up your program! I am not a software designer, although I have a son who is, but these days commercial software is available that lets you get farther down the line of designing steps that will allow you to measure features of what is seen in the cell. These features are identified by their brightness, their shape, their nearness to other objects, and so on. Once a strategy is developed, you can sit back and analyze every image in a directory, watch images whiz by on the screen while the data is dumped into an Excel spreadsheet. It is a totally energizing exercise because it represents a finish to a tedious slog to get the program/ journal to work. This DNA project is in the stage of method development. The result will be a proof of principle readying them for the ultimate: clinical experiments. More work, more work, in quarantine, but the team will soon/eventually go back to the lab. I remain here in Western North Carolina virtually ready to help.
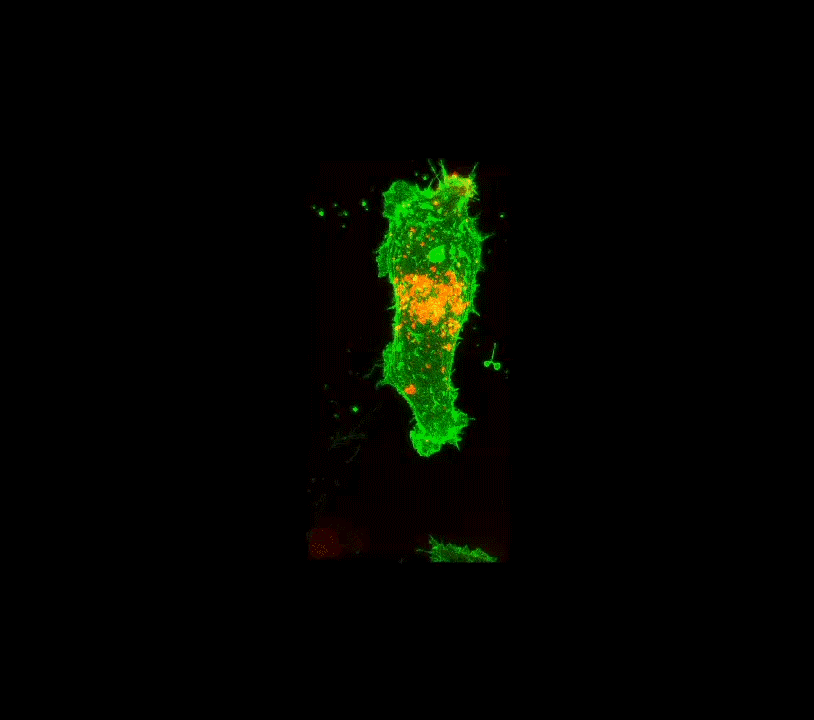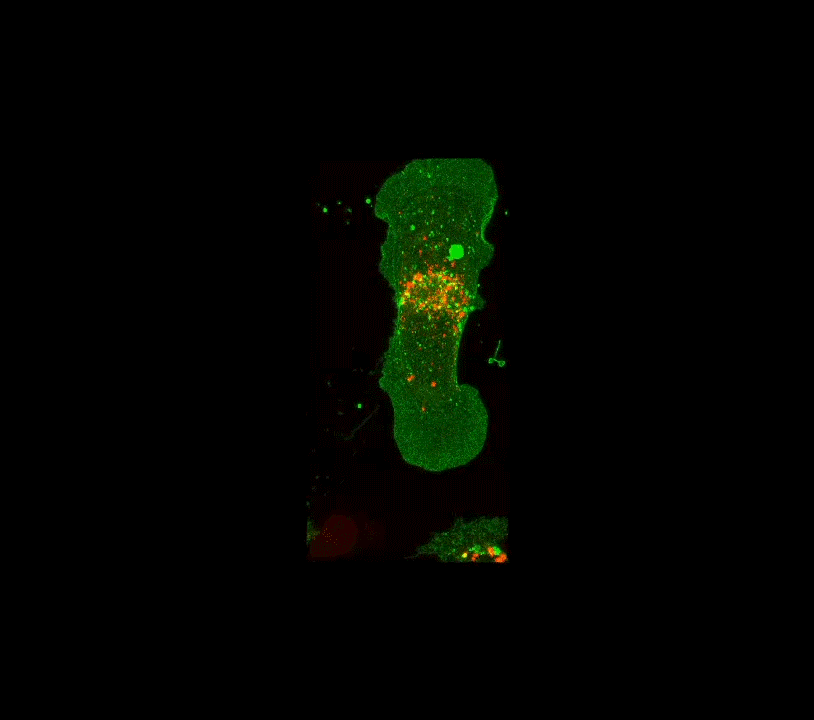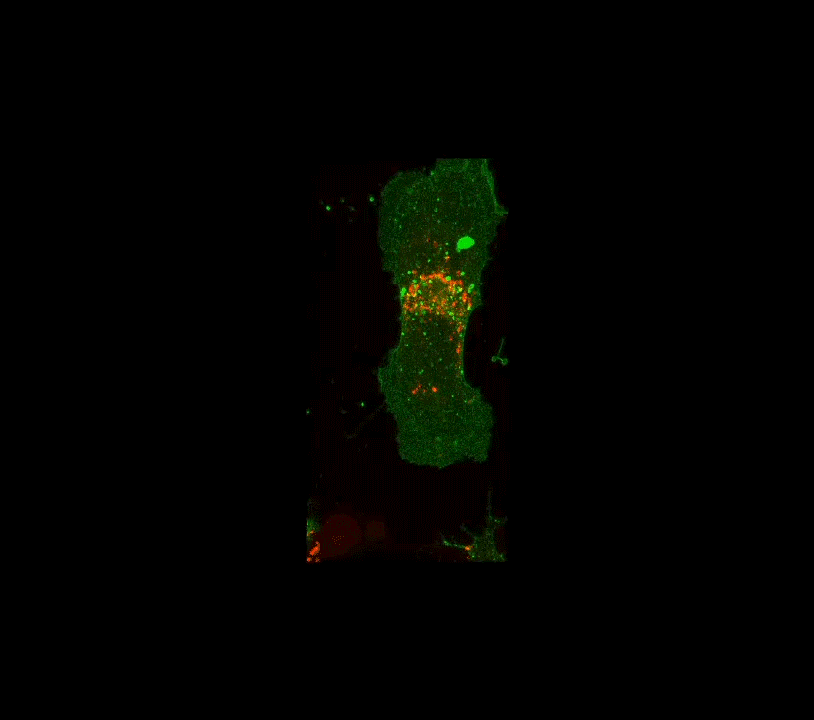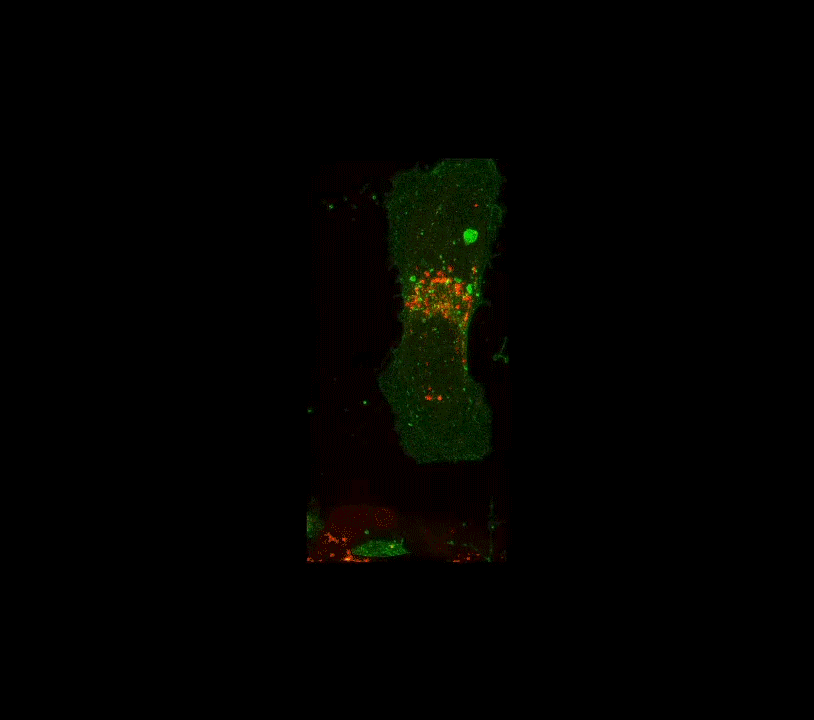
Along these lines, here is an example below of work done a few years ago when I was still part time and traveling to D.C.: confocal images of the interior of an individual cell (in black and white, but the fluorescence is actually green!). My colleague, her PhD student and I were studying how the angiotensin receptor moves within the cell when the cell is confronted with angiotensin. These two molecules together regulate blood pressure. The model system is very simple, it consists of human cells cultured on glass substrates within an imaging chamber.
4 confocal microscope images taken over 20 minutes time. A single living cell and part of an adjacent linked cell is observed in living cells. One particular protein receptor was molecularly linked to green fluorescent protein. Within 5 minutes after adding angiotensin, its fluorescent receptor, normally at the cell surface, is taken into the cell into tiny (white) membrane vesicles.
I need to show you a pretty green fluorescent image as well, especially if you didn’t go to the Leica link above! This cell has two colors to ask the question does the green protein go to places in the cell where the red is (lysosome)? It is a live movie that in total real time would be 25 min I recall. It is a pretty movie that is a visual representation of data (number of photons captured over time in every voxel or cube of 3D space). Unfortunately this pretty movie has several technical problems with the data. I don’t need to go into that here!! But, this early test of the methods was the basis of subsequent experiments. You would love the names of the parts of the cell surface that is so dynamically displayed here: lamellipodium, ruffling, internalization, vesicles, and so on. I love this stuff.
3D movie of one cell expressing green fluorescent receptor and red-labeled lysosomes (where proteins are “digested” within the cell). Individual images were taken in a series of vertical stacks covering the height of the cell every 1 minute for about 25 minutes. 25 3D images were the result after computer reconstruction.
We had to work out not only the details of imaging but also the image analysis methods to measure the movement of the fluorescent receptor from the cell surface to the cell interior. Finally, after much work, the student and my collaborators together with me published a methods paper. It was published in the journal Journal of Visualized Experiments (JoVE). This journal is rather unique in that together with the authors JoVE provides a video version demonstration of the methods together with the written detailed instructions. This is what the citation looks like:
Live Cell Imaging and 3D Analysis of Angiotensin Receptor Type 1a Trafficking in Transfected Human Embryonic Kidney Cells Using Confocal Microscopy. J Vis Exp. 2017 Mar 27;(121):55177.
Parnika Kadam 1, Ryan McAllister 2, Jeffrey S Urbach 2, Kathryn Sandberg 1, Susette C Mueller 3
1 Department of Biochemistry, Georgetown University Medical Center; Department of Medicine, Georgetown University Medical Center.
2 Department of Physics, Georgetown University Medical Center.
3 Department of Oncology, Georgetown University Medical Center; muellers@georgetown.edu.
How did this bit of science influence my pieces? Here is the link to my Instagram showing a t-shirt I made worn by the PhD student Parnika Kadam who performed this work. I did a wall hanging for the office of by colleague/collaborator Kathryn Sandberg, PhD.
The next blog will be about the tree silkscreen and next steps of the project that I started to detail from start to finish a hand dyed and quilted wall hanging. I promise!